
Research
In our laboratory, we are developing chemical biology techniques to label and visualize proteins with synthetic fluorescent molecules by devising and applying chemical principles. In living cells, countless biomolecules exist, dynamically changing their localization and controlling cellular events by performing the biomolecular functions in a subcellular region where they are needed. Visualization of the movement of these biomolecules provides important information to elucidate the physiological functions they control. We have developed original technology for fluorescent labeling of proteins to reveal how proteins move in living cells and regulate biological phenomena. Furthermore, we aim to elucidate biological phenomena regulated by nucleic acids, glycans, and extracellular vesicles in addition to proteins, and to control functions of biomolecules at will by making full use of our protein labeling technology.
Development of protein labeling technology using PYP tag and synthetic fluorescent probes
Protein labeling using protein tags and synthetic fluorescent probes has recently been developed as a new chemistry-based imaging technique. In this technique, a protein tag fused to a protein of interest (POI) is specifically labeled with a synthetic fluorescent probe to visualize dynamics of POI. This technology enables highly accurate spatio-temporal analysis of protein dynamics by pulse-chase experiments and has also been applied to super-resolution imaging.
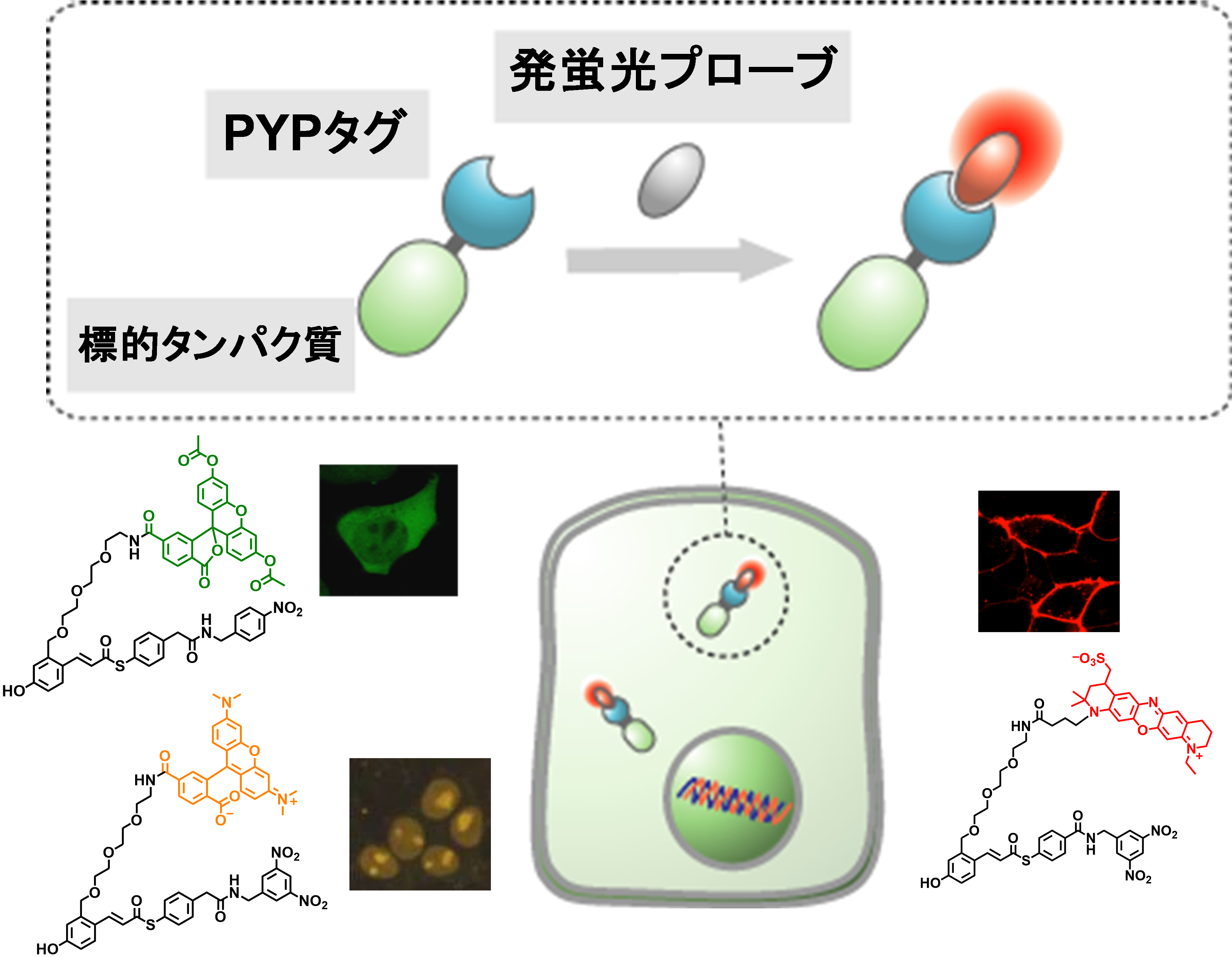
On the other hand, since conventional probes always emit fluorescence, fluorescence of free probes generates background signals when cells are incubated with probes and imaged using a microscope without cell washing, compromising detection of POIs. To solve this problem, we have developed a protein labeling technology that uses an original protein tag called PYP tag and its labeling probes that become fluorescent upon labeling reaction. By applying this technology, we have visualized dynamics of various proteins involved in epigenetics, immune responses, and tumorigenesis. Currently, we are developing fluorescent probes for faster and higher contrast protein imaging.
【References】1. J. Am. Chem. Soc. 2009, 131, 16610. / 2. Angew. Chem. Int. Ed. 2012, 51, 5611. / 3. J. Am. Chem. Soc. 2013, 135, 12360. / 4. Angew. Chem. Int. Ed. 2015, 54, 14368. / 5. Chem. Sci. 2016, 7, 308. / 6. Chem. Sci. 2020, 11, 3694–3701.
Elucidation of GLUT4 dynamics by protein labeling technology
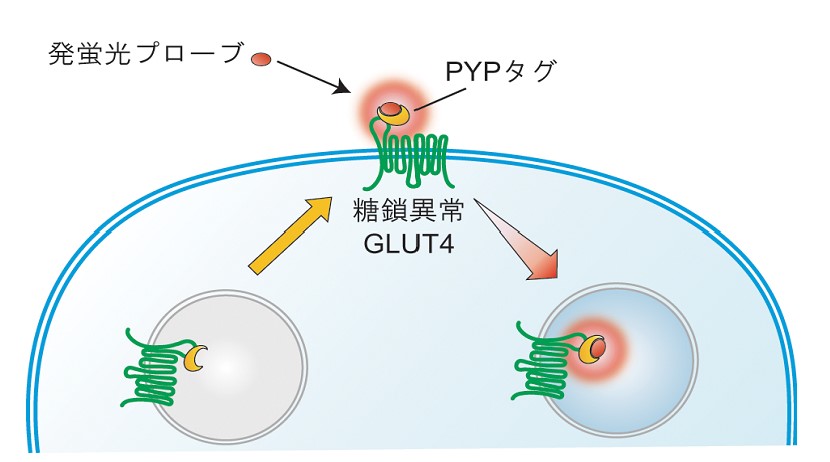
GLUT4 belongs to glucose transporter family and plays an important role in reduction of blood glucose levels by taking up glucose into cells. Patients with type 2 diabetes have a problem in GLUT4 translocation to the plasma membrane and cannot efficiently take up glucose into cells. Therefore, elucidation of the dynamic regulation mechanism of GLUT4 is an extremely important issue for elucidating the pathogenesis and treatment of diabetes. Recently, the role of glycans bound to GLUT4 in the intracellular dynamics of GLUT4 has attracted much attention. Using protein labeling technology, we have visualized translocation of GLUT4 to the plasma membrane and found that glycans play a role in retaining GLUT4 at the plasma membrane. We are currently conducting research to elucidate the details of the mechanism by which glycans regulate the dynamics of GLUT4 in order to gain knowledge that will be useful for the treatment of diabetes.
【References】1. Nat. Chem. Biol. 2016, 12, 853. / 2. Chem. Lett. 2020, 49, 232.
Development of fluorescent probes to visualize protein degradation
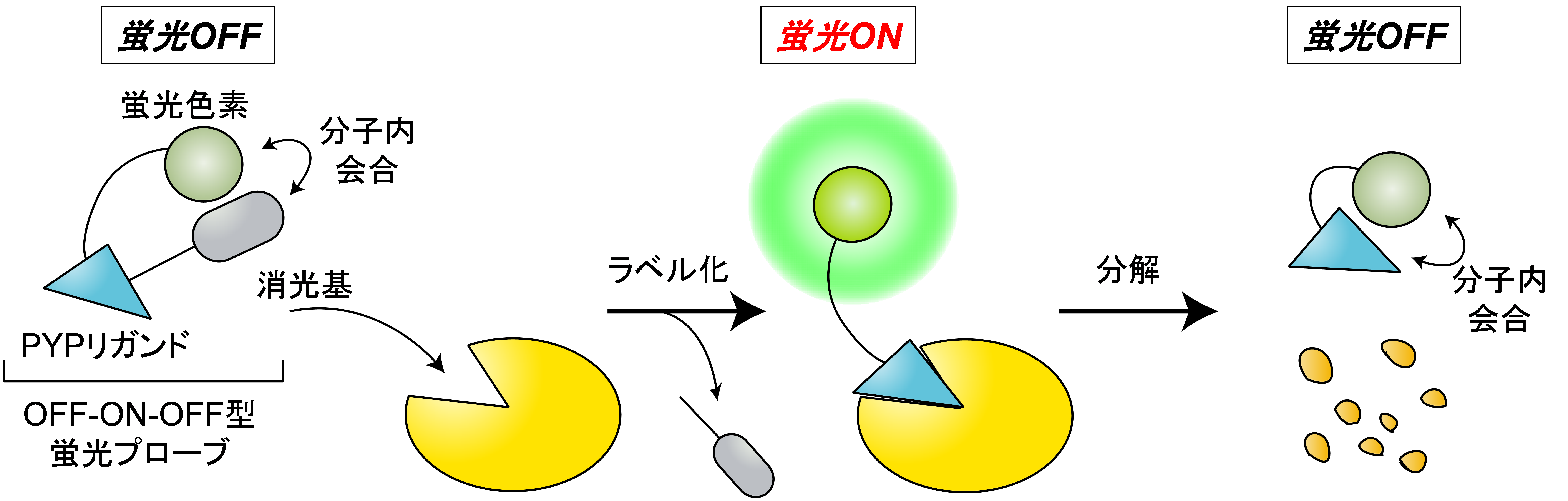
Protein degradation plays a pivotal role in cell signaling and cell cycle regulation, in addition to maintaining the nutritional environment and removing abnormal proteins. Abnormalities in the balance between protein expression and degradation are known to cause cancer and neurological diseases. In addition, compounds that artificially induce protein degradation are attracting great attention as new pharmaceuticals. Therefore, visualization of protein degradation in living cells is expected to provide useful information on the mechanism of protein degradation and to be a new tool for medicine and drug discovery. We have succeeded in visualizing protein degradation by developing an "OFF-ON-OFF type fluorescent probe," which is non-fluorescent in the free state, becomes fluorescent when a POI is labeled, and decreases in fluorescence intensity again when the POI is degraded. Currently, we are developing technology to detect protein degradation with even higher sensitivity.
【References】1. Bioconjug. Chem. 2020, 31, 577–583. / 2. Chem. Sci. 2022, 13, 1419.
Visualization of nucleic acid modifications by hybrid probes
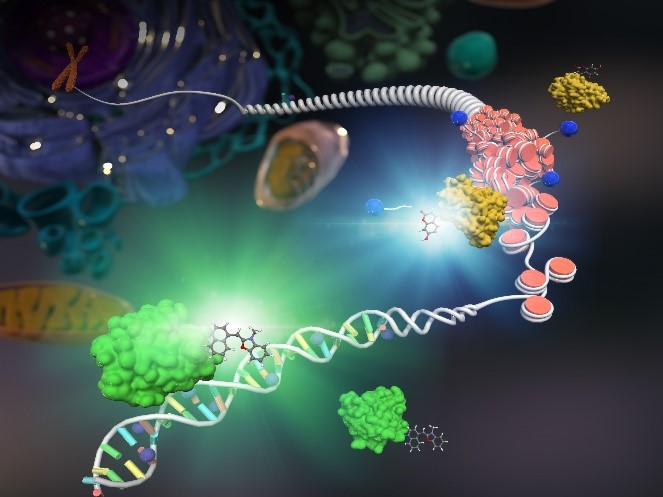
Epigenetics is a mechanism to control gene expression without alteration of DNA sequences, and has been attracting a great deal of attention in recent years. In particular, DNA methylation is one of representative chemical modifications in the epigenetic mechanism, and its abnormalities are known to cause various cancers and immune diseases. For this reason, elucidation of the mechanism is of great importance not only in life sciences but also in the fields of medicine and drug discovery. So far, many biochemical methods for analyzing methylated DNA with cell lysis have been reported. On the other hand, the development of methods to analyze DNA methylation in living cells would be extremely useful to elucidate the dynamics of DNA methylation, but these methods are limited to fluorescent protein-based methods. We have applied protein labeling technology to construct hybrid probes consisting of synthetic dyes and proteins in cells. The hybrid probe binds specifically to methylated DNA and increases the fluorescence intensity. With this hybrid probe, we have succeeded in visualizing the dynamics of methylated DNA in living cells. While various chemical modifications of nucleic acids other than DNA methylation are known, there are many issues to be elucidated regarding their intracellular dynamics and functions. For this reason, we are now working on the development of hybrid probe technology to visualize the chemical modifications of those nucleic acids and to elucidate their functions.
【References】1. J. Am. Chem. Soc. 2018, 140, 1686. / 2. Acc. Chem. Res. 2019, 52, 2849–2857.
Development of photoswitchable fluorescent probes for super-resolution imaging
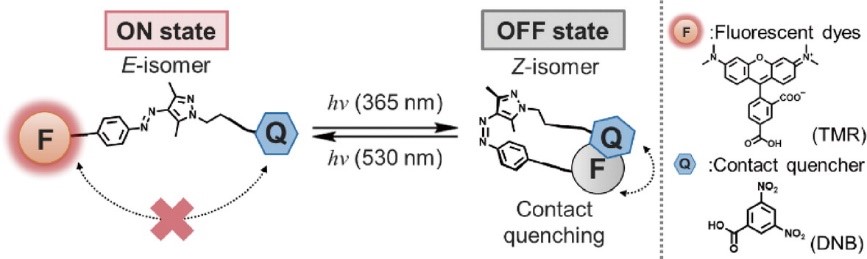
Photoswitchable fluorescent probes are functional molecules, which turn off and on the fluorescence intensity upon light, irradiation, and are powerful tools for visualizing the dynamics of biomolecules in super-resolution microscopy. In particular, there is a great need for molecules that can control fluorescence intensity only with visible light without photofatigue or photobleaching, enabling long-term super-resolution imaging in living cells. On the other hand, photoswitchable fluorescent probes have been developed by modifying fluorescent proteins and synthetic fluorophores, but their susceptibility to photofatigue or photobleaching and cytotoxicity have limited the development of live-cell super-resolution technology. We are developing photoswitchable fluorescent probes based on a new concept by utilizing and modifying photochromic molecules to solve this problem.
【References】Bull. Chem. Soc. Jpn. 2022, 93, 821.
page top
